Extract DataFrame from Compressed Data into Python Datatable#
Compressed Data Extraction in Python Datatable#
In a previous blog post, I discussed the various ways to extract data from compressed files into Pandas. This blog post explores those options, this time with Datatable. This package is a reimplementation in Python of Rdatatable, aiming to replicate its speed and concise syntax. Have a look at my other blog post that explores the basics of Datatable.
Most of the examples will focus on the zip format, as it is widely used and the methods presented here can be adapted for other compression formats.
Read Zip Folder that has only one File#
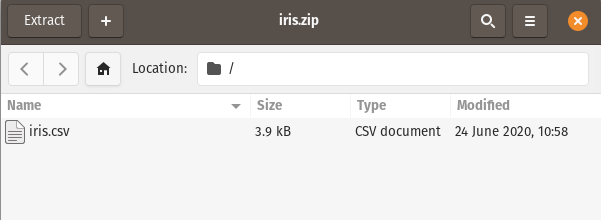
We simply pass the zipfile :
from datatable import dt, fread, iread
df = fread("Data_files/iris.zip")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
**Notes: **
Read a File from Multiple Files in Zip Folder#
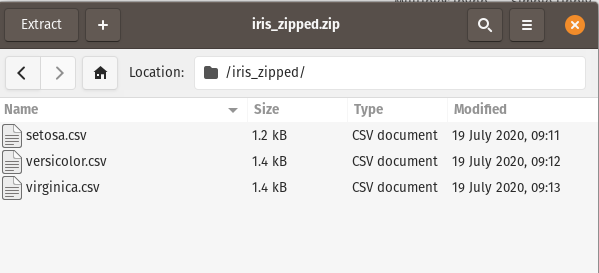
In this scenario, We want just the setosa.csv file. Datatable can read this with ease - simply pass the filepath to fread -> archive_name.zip/path_to_file:
df = fread("Data_files/iris_zipped.zip/iris_zipped/setosa.csv")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
This is easier than working with Pandas, where we had to import the zipfile to do this.
Read a Specific File from Multiple Files in Zip Folder based on its Extension#
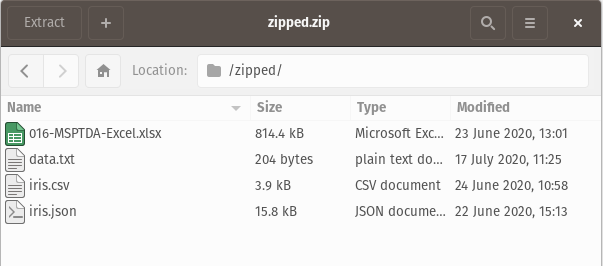
Here, the zip folder contains various file types (xlsx, csv, json, …), and we want only the .csv file. Let’s also assume that we do not know beforehand what the filename is; we do however know that there is just one .csv file in the zip folder. There are two options :
Option 1 : We can pick out the specific file with the iread function. iread returns a lazy iterator of all the data frames in the zip folder; as such we can filter for the specific file we are interested in :
for file in iread("Data_files/zipped.zip"):
# every file has a source attribute
# that describes the file,
# including the file type and name
if file.source.endswith(".csv"):
df = file
df.head()
IOWarning: Could not read Data_files/zipped.zip/zipped/iris.json: Too few fields on line 151: expected 6 but found only 5 (with sep=','). Set fill=True to ignore this error. << {"sepalLength": 5.9, "sepalWidth": 3.0, "petalLength": 5.1, "petalWidth": 1.8, "species": "virginica"}>>
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Note:
The
.jsonformat is not supported by Datatable.
Option 2 : Use the cmd argument. This passes the processing to the command line. The command line is a powerful tool that can help with a lot of data processing. No need for the iread function, just let the command line do the work.
df = fread(cmd = """unzip -p Data_files/zipped.zip '*.csv' """)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Note :
In the code above, we used the unzip tool to filter for the
csvfile and extract the data.
Read Multiple Files with specific extension in Zip Folder into a Single Dataframe#
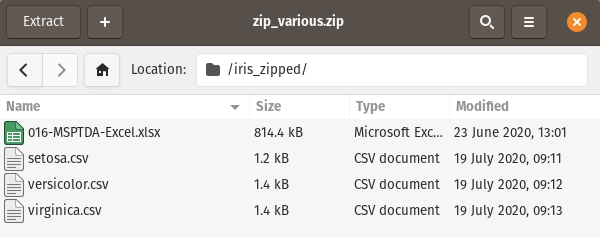
In this scenario, we want to read in only the csv files and combine them into one dataframe - We will explore the iread option and the command line option:
Option 1 - iread :
csv_files = [file for file in iread("Data_files/zip_various.zip")
if file.source.endswith(".csv")]
# combine the dataframes into one with rbind
# rbind is similar to pd.concat(*frames, axis=0) :
df = dt.rbind(csv_files)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Option 2 - Command Line :
df = fread(cmd = """
unzip -p Data_files/zip_various.zip '*.csv' |
awk 'BEGIN {FS = ","}; {if ($1!="sepal_length" || NR==1) {print}}'
""")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Note:
The AWK language is used here to split the columns on the
,delimiter, and to get the header for the rows.
Read all the Files in Zip Folder into a Single Dataframe#

Option 1 - iread :
#get a sequence of the files and combine with rbind
df = dt.rbind(tuple(iread("Data_files/iris_zipped.zip")))
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Option 2 - Command Line :
df = fread(cmd = """
unzip -p Data_files/iris_zipped.zip |
awk 'BEGIN {FS = ","}; {if ($1!="sepal_length" || NR==1) {print}}'
""")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
What about other Formats?#
The major compression archive formats supported by fread are : .tar, .gz, .zip, .gz2, and .tgz. So the above steps will work for these formats. But there are other compresssion formats, including .7z. How do we read that? The command line!
Extract archive with a single file :
#ensure 7z is already installed on your system
df = fread(cmd = "7z e -so Data_files/iris_single_7z.7z")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
Extract specific file :
df = fread(cmd = """ 7z e -so Data_files/iris_7z.7z "virginica.csv" -r """)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪▪▪▪▪ | ▪▪▪▪ | |
| 0 | 6.3 | 3.3 | 6 | 2.5 | virginica |
| 1 | 5.8 | 2.7 | 5.1 | 1.9 | virginica |
| 2 | 7.1 | 3 | 5.9 | 2.1 | virginica |
| 3 | 6.3 | 2.9 | 5.6 | 1.8 | virginica |
| 4 | 6.5 | 3 | 5.8 | 2.2 | virginica |
| 5 | 7.6 | 3 | 6.6 | 2.1 | virginica |
| 6 | 4.9 | 2.5 | 4.5 | 1.7 | virginica |
| 7 | 7.3 | 2.9 | 6.3 | 1.8 | virginica |
| 8 | 6.7 | 2.5 | 5.8 | 1.8 | virginica |
| 9 | 7.2 | 3.6 | 6.1 | 2.5 | virginica |
Summary#
Datatable offers convenient features in reading data from compressed file formats, including the ability to read in data via the command line. Compared to Pandas, Datatable offers more flexibility and power when dealing with compressed file formats.
Comments#