Extract DataFrame from Compressed Data into Pandas#
Compressed Data Extraction in Pandas#
You have data in compressed form (zip, 7z, …); how do you read the data into Pandas? This blog post will explore the various options possible. The focus will be on the zip format, as it is widely used and the methods presented here can be adapted for other compression formats.
Read Zip Folder that has only one File#
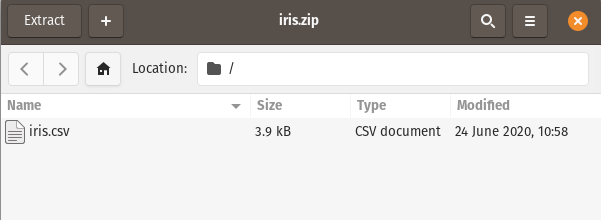
Pandas can easily read a zip folder that contains only one file:
import pandas as pd
df = pd.read_csv("Data_files/iris.zip")
# or : pd.read_csv("Data_files/iris.zip",
# compression = "zip")
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Note:
Note that Pandas successfully inferred the compression type. It pays sometimes though, to be explicit and supply the
compressionargument.
Read a File from Multiple Files in Zip Folder#
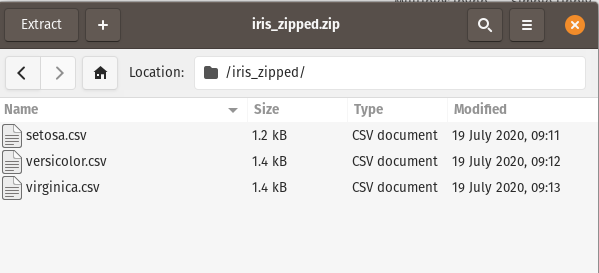
In this scenario, We want just the setosa.csv file. Pandas cannot directly read data from a zip folder if there are multiple files; to solve this, we will use the zipfile module within Python. The zipfile module offers two routes for reading in zip data : ZipFile and Path classes.
Option 1 - Via ZipFile :
from zipfile import ZipFile, Path
# pass in the specific file name
# to the open method
with ZipFile("Data_files/iris_zipped.zip") as myzip:
data = myzip.open("iris_zipped/setosa.csv")
#Now, we can read in the data
df = pd.read_csv(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Note:
The context manager ensures the connection is closed after reading in the data.
The complete path to the specific file is passed (
iris_zipped/setosa.csv).
Option 2 - Via Path:
from io import StringIO
# the 'at' argument accepts the path to the file
# within the zip archive
# this is convenient when you know the filename
zipped = Path("Data_files/iris_zipped.zip", at="iris_zipped/setosa.csv")
# read into dataframe, with the '.read_text' method
df = pd.read_csv(StringIO(zipped.read_text()))
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Read a Specific File from Multiple Files in Zip Folder based on its Extension#
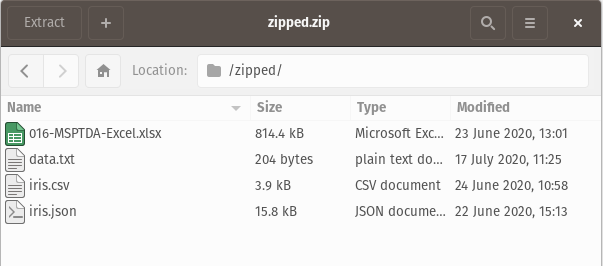
Here, the zip folder contains various file types (xlsx, csv, json, …), and we want only the .csv file. Let’s also assume that we do not know beforehand what the filename is; we do however know that there is just one .csv file in the zip folder. We can pick out the specific file with the zipfile module:
Option 1 - Via ZipFile :
#fnmatch helps to filter for specific file types
import fnmatch
#read data from the csv file
with ZipFile("Data_files/zipped.zip") as zipped_files:
#get list of files in zip
file_list = zipped_files.namelist()
#use fnmatch.filter to get the csv file
csv_file = fnmatch.filter(file_list, "*.csv")
#get the csv data
data = zipped_files.open(*csv_file)
#read into dataframe
df = pd.read_csv(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Option 2 - Via Path:
zipped = Path("Data_files/zipped.zip")
#filter for csv file
csv_file = fnmatch.filter(zipped.root.namelist(), "*.csv")
#append filepath to zipped, and read into dataframe
data = zipped.joinpath(*csv_file).read_text()
df = pd.read_csv(StringIO(data))
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Read Multiple Files with specific extension in Zip Folder into a Single Dataframe#
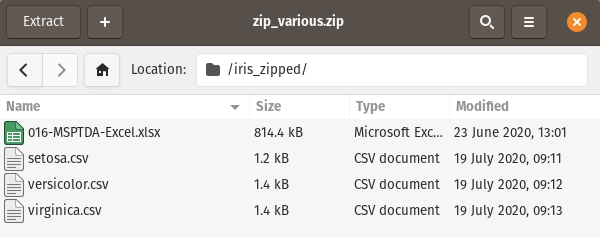
In this scenario, we want to read in only the csv files and combine them into one dataframe - zipfile module to the rescue:
Option 1 - Via ZipFile:
with ZipFile("Data_files/zip_various.zip") as zipfiles:
file_list = zipfiles.namelist()
#get only the csv files
csv_files = fnmatch.filter(file_list, "*.csv")
#iterate with a list comprehension to get the individual dataframes
data = [pd.read_csv(zipfiles.open(file_name)) for file_name in csv_files]
#combine into one dataframe
df = pd.concat(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Option 2 - Via Path:
zipped = Path("Data_files/zip_various.zip")
csv_file = fnmatch.filter(zipped.root.namelist(), "*.csv")
data = (zipped.joinpath(file_name).read_text() for file_name in csv_file)
data = (pd.read_csv(StringIO(content)) for content in data)
df = pd.concat(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Read all the Files in Zip Folder into a Single Dataframe#

Option 1 - Via ZipFile:
with ZipFile("Data_files/iris_zipped.zip") as zipfiles:
#the first entry is the zipfile name
#we'll skip it
filelist = zipfiles.namelist()[1:]
data = [pd.read_csv(zipfiles.open(file_name)) for file_name in filelist]
df = pd.concat(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Option 2 - Via Path:
zipped = Path("Data_files/iris_zipped.zip")
data = (zipped.joinpath(file_name).read_text()
for file_name in zipped.root.namelist()[1:])
data = (pd.read_csv(StringIO(content)) for content in data)
df = pd.concat(data)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Read Compressed Data through the Command Line#
This process is especially useful when dealing with large files that Pandas may struggle with. Preprocess the data, before reading it into Pandas. The command line tools are quite powerful and allow for some efficient data processing.
import subprocess
#read a zip with only one file
data = subprocess.run("unzip -p Data_files/iris.zip",
shell = True,
capture_output = True,
text = True).stdout
#read in data to pandas
df = pd.read_csv(StringIO(data))
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Note:
The unzip tool on the command line does the hardwork here.
# filter for only rows where species == virginica
# this comes in handy when dealing with large files
# that Pandas may struggle with
data = subprocess.run("""
unzip -p Data_files/iris.zip | awk 'BEGIN {FS = ","};
{if($5=="virginica" || NR == 1) {print}}'
""",
shell = True,
capture_output = True,
text = True).stdout
df = pd.read_csv(StringIO(data))
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 6.3 | 3.3 | 6.0 | 2.5 | virginica |
| 1 | 5.8 | 2.7 | 5.1 | 1.9 | virginica |
| 2 | 7.1 | 3.0 | 5.9 | 2.1 | virginica |
| 3 | 6.3 | 2.9 | 5.6 | 1.8 | virginica |
| 4 | 6.5 | 3.0 | 5.8 | 2.2 | virginica |
Note:
The AWK language is used here to filter for the rows and also retain the headers.
Reading via the command line can be simplified with pyjanitor’s read_commandline function:
# pip install pyjanitor
from janitor import read_commandline
#read a zip with only one file
cmd = "unzip -p Data_files/iris.zip"
df = read_commandline(cmd = cmd)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
cmd = "unzip -p Data_files/iris.zip |"
cmd += """awk 'BEGIN {FS = ","}; """
cmd += """{if($5=="virginica" || NR == 1) {print}}'"""
df = read_commandline(cmd = cmd)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 6.3 | 3.3 | 6.0 | 2.5 | virginica |
| 1 | 5.8 | 2.7 | 5.1 | 1.9 | virginica |
| 2 | 7.1 | 3.0 | 5.9 | 2.1 | virginica |
| 3 | 6.3 | 2.9 | 5.6 | 1.8 | virginica |
| 4 | 6.5 | 3.0 | 5.8 | 2.2 | virginica |
Summary#
This blog post showed various ways to extract data from compressed files. Personally, I feel extraction using the ZipFile function is simpler and cleaner than the Path function. Also, we saw the power of the command line and how useful it can be. In a future blog post, I will explore the command line in more detail.
Comments#